

Flexible cost
For many people, health insurance covers the cost of genetic testing. Invitae also offers financial assistance for those who qualify.





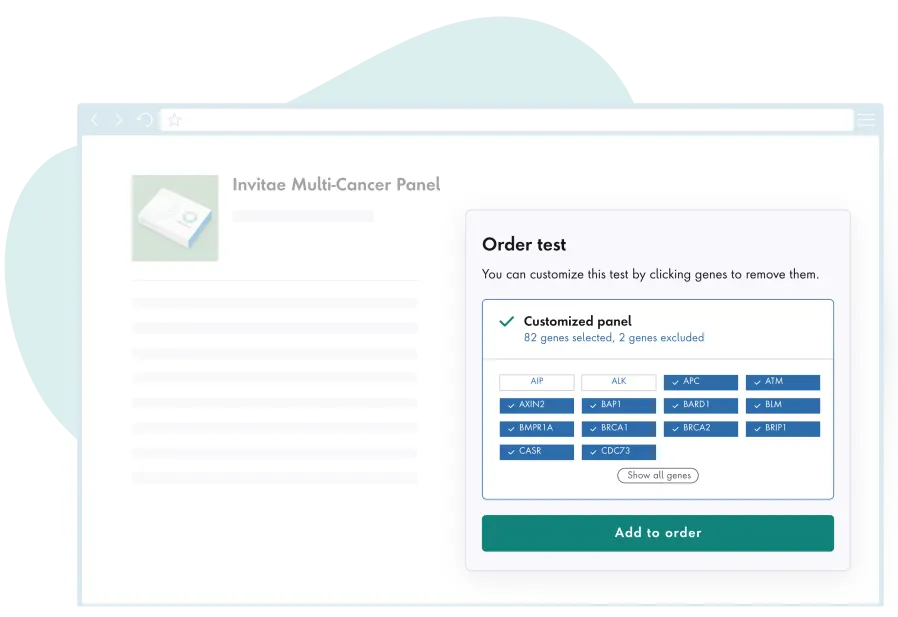
Actionable insights
Invitae's online portal and genetic counseling services can help interpret test results.
Learn moreInvitae genetic testing in three easy steps
Step 1
Start the ordering process via our convenient online portal. A doctor will place an order for you.
Step 2
Collect a specimen using an Invitae collection kit and send it back to us.
Step 3
Receive results online. Genetics experts can help answer questions.
What's new
Driving the field of genetics forward, one announcement at a time.
Invitae Generation™️
It’s an evolution of technology—powered by people. Learn more about our genetic testing and classification methods.
Universal testing?
A recent study suggests current genetic testing guidelines may miss patients.
EHR integration
Explore the potential of electronic health record (EHR) integration in simplifying genetic testing workflows.
What's a genetic counselor...
...and why do I need one? Find out in this blog post.
Let's get started
Interested in talking with Invitae about genetic testing in more detail? Schedule a 5 minute call.
If you are a patient, please email us at clientservices@invitae.com.
Have questions?
- How are Invitae’s genetic tests different from other genetic tests?
Invitae’s broad test offerings inform every stage of life for patients and their families, providing a single, reliable source for medical-grade genetic testing. Tests come with flexible billing options and built-in support to make confident health decisions based on results.
- Are genetic counseling services available to all patients undergoing Invitae testing?
A genetic counseling session is available to answer brief questions about genetic testing and results. This is included at no additional cost to patients located in the US, US territories, and Canada. The only tests that do not qualify are pharmacogenomics screening or personalized cancer monitoring. Learn more on the consult with a genetic expert page.
- What payment options are available?
Through the Invitae Billing Assurance Program, our dedicated billing specialists work with each patient to get a comprehensive view of their individual situation and help identify cost-effective and affordable payment options. Billing insurance is often the most cost-effective choice. Invitae is in network with national US health insurance plans, covering more than 300 million patients in the United States. Typically, the out-of-pocket cost for a patient using insurance is less than $100. Invitae also accepts HSA/FSA payments.
- Can I contact Invitae today?
Yes, you can contact Invitae today by visiting our contact us page to find contact information for client services and billing specialists. To speak directly with client services, email clientservices@invitae.com at any time or call 800-436-3037 Monday through Friday, 5:00 am to 5:00 pm Pacific time.